Protein interactions serve as the cornerstone of functionality, yet their complexity often defies easy characterization. Mass photometry is a tool that can shed light on these intricate dynamics. By directly measuring the molecular mass of individual biomolecules in solution, mass photometry accurately identifies binding partners, complexes formed, and the strength of their affinities.
In this white paper, explore a recent study that uses mass photometry to shed light on how human and bovine Immunoglobulin G (IgG) antibodies interact with protein A. By revealing distinct variations in complex formation and binding strengths, this label-free technique offers a deeper understanding of critical biological mechanisms.
Discover more about how mass photometry resolves complex equilibria, ensuring accurate quantification and assessment of protein-protein interactions. From unraveling intricate binding affinities to assessing sample homogeneity and purity, this technology is pivotal to understanding how proteins function together, and are affected by different conditions.
Download this whitepaper to learn:
- The intricacies of protein-protein interactions and how these can be quantified using mass photometry
- How mass photometry resolves complex equilibria, aiding in precise quantification
- Practice applications and implications of protein binding affinity quantifications in research and biotechnology
Antibody binding affinities application note
®
APPLICATION NOTE
Quantifying protein binding affinities using mass photometry
Mass photometry is a label-free bioanalytical tool that can assess the dynamics and strength of protein-protein interactions by directly measuring the mass and the relative abundance of individual proteins as well as the complexes they form in solution.
Protein interactions play pivotal roles in a wide range of biological processes. However, they are often highly complex, involving the binding of multivalent ligands to multiple binding sites. As a result, it can be challenging to identify the various complexes formed and assess the strength of their interactions.1
Mass photometry is a fast, label-free technology that measures the molecular mass of individual biomolecules in solution, and is ideal for studying protein-protein interactions. By providing the mass distribution of biomolecules in a sample, it gives a detailed overview of the binding partners present, the complexes they form, the relative abundance of each species and the strength of their interactions.1,2,3
Here, we apply mass photometry to characterize interactions between Immunoglobulin G (IgG) antibodies of different origin species (human and bovine) and protein A. We quantify the relative abundance of each protein and the complexes they form in solution. We use the automated pipetting feature of Refeyn’s TwoMP Auto mass photometer to generate highly reproducible data. From these measurements, we calculate the equilibrium dissociation constant (KD) for each interaction.
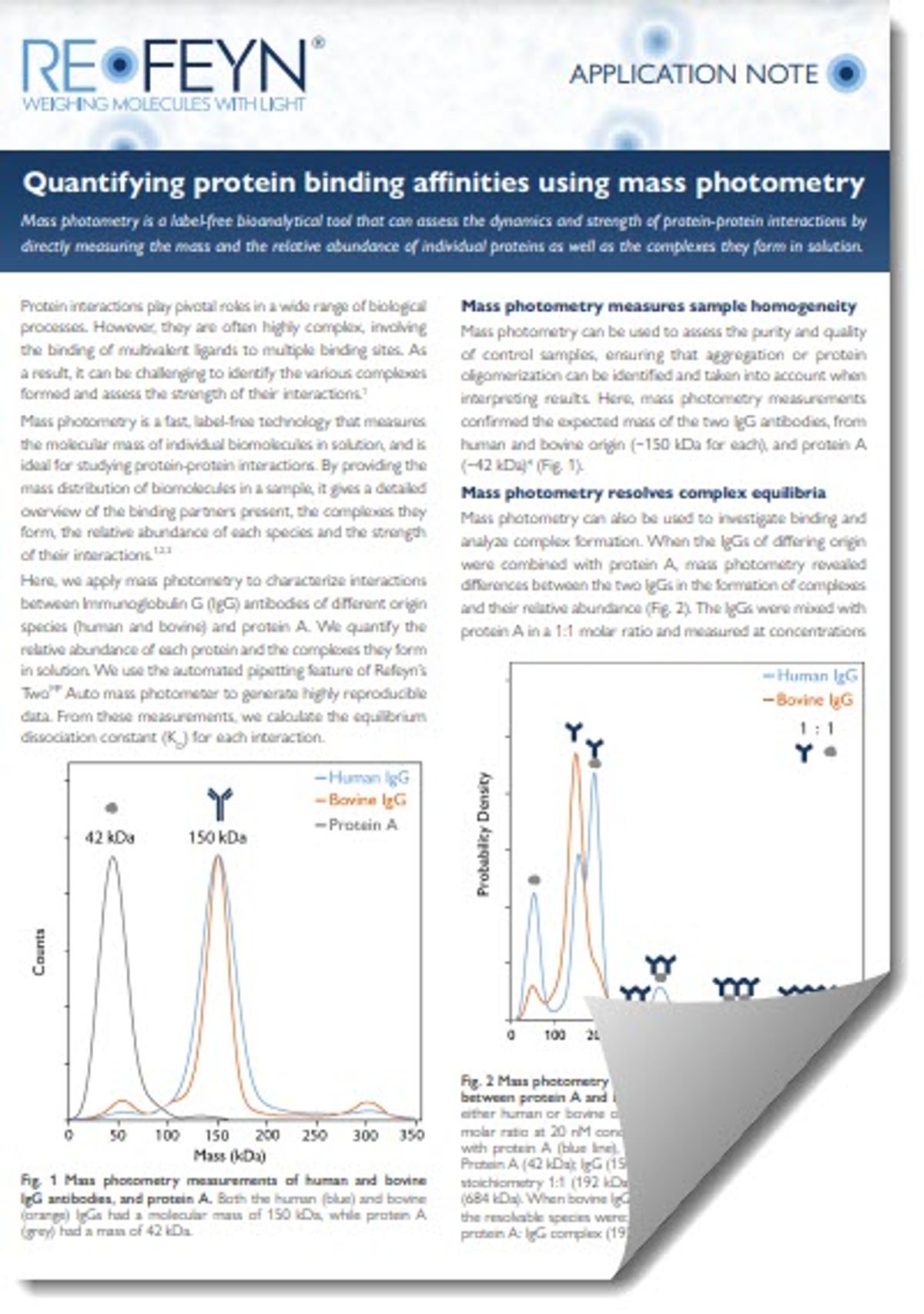
Fig. 1 Mass photometry measurements of human and bovine IgG antibodies, and protein A. Both the human (blue) and bovine (orange) IgGs had a molecular mass of 150 kDa, while protein A (grey) had a mass of 42 kDa.
Mass photometry measures sample homogeneity
Mass photometry can be used to assess the purity and quality of control samples, ensuring that aggregation or protein oligomerization can be identified and taken into account when interpreting results. Here, mass photometry measurements confirmed the expected mass of the two IgG antibodies, from human and bovine origin (~150 kDa for each), and protein A (~42 kDa)4 (Fig. 1).
Mass photometry resolves complex equilibria
Mass photometry can also be used to investigate binding and analyze complex formation. When the IgGs of differing origin were combined with protein A, mass photometry revealed differences between the two IgGs in the formation of complexes and their relative abundance (Fig. 2). The IgGs were mixed with protein A in a 1:1 molar ratio and measured at concentrations
Fig. 2 Mass photometry reveals differences in complex formation between protein A and IgG antibodies of differing origin. IgGs of either human or bovine origin were mixed with protein A in a 1:1 molar ratio at 20 nM concentration. When human IgG was mixed with protein A (blue line), the following species could be resolved: Protein A (42 kDa); IgG (150 kDa); and protein A: IgG complexes of stoichiometry 1:1 (192 kDa), 1:2 (342 kDa), 2:3 (534 kDa) and 2:4 (684 kDa). When bovine IgG was mixed with protein A (orange line), the resolvable species were: Protein A (42 kDa), IgG (150 kDa), 1:1 protein A: IgG complex (192 kDa) and IgG dimers (300 kDa).
of 5, 10 and 20 nM. At all concentrations measured, both IgGs formed 1:1 complexes with protein A, but differences emerged for higher-order complexes.
Protein A interacted strongly with human IgG, as demonstrated by the formation of higher-order protein A:IgG heterocomplexes and the low percentage of free human IgG (33%). In contrast, protein A bound less readily to bovine IgG. In the case of bovine IgG, higher-order protein A:IgG complexes were not observed, and the majority of bovine IgG remained unbound (82%). The results show that mass photometry can resolve and quantify the relative abundance of the multiple species that co-exist in complex equilibrium reactions.
Mass photometry measures interaction strength
We observed above that protein A forms more complexes and higher-order complexes with the human IgG antibody as compared to the bovine IgG. This indicates a difference in affinity between the two interactions
To quantify this difference, we calculated the KD for each interaction (Fig. 3) from mass photometry measurements at 20 nM concentration (the data shown in Fig. 2 and two repeats). The calculations confirmed that protein A binds to the bovine
Fig. 3 Multiple Kd values can be calculated from a single mass photometry measurement. Schematics show the complex formation depicted in Fig. 2. KD values for interactions involving the
IgG of human origin (blue) and of bovine origin (orange) are both
shown. Since IgGs of bovine vs. human origin did not form all the same complexes, the KD calculation was not applicable for certain interactions (depicted as N/A). The KD values were calculated for
each of three repeated mass photometry measurements, and the
mean ± standard deviation is given.
IgG with lower affinity (KD = 69.4 ± 9.0 nM) than it binds to the human IgG (KD = 5.73 ± 2.6 nM). In addition, for the human IgG – protein A interaction, slightly lower KD values were observed for larger complexes, suggesting possible cooperativity effects involving IgG and/or protein A.
While we repeated the experiment to improve the accuracy of the measurement, a single mass photometry measurement can provide the data needed to calculate KD, even for complex equilibria. Mass photometry can be used to measure KD values for any interactions where both bound and unbound species are observable at the concentrations used in mass photometry measurements.
Conclusion
Mass photometry is a versatile, label-free bioanalytical tool that can be used to investigate complex equilibria at physiologically relevant concentrations. It can rapidly measure the mass of individual biomolecules and the complexes they form in solution, using very little sample.
A single mass photometry measurement can generate the data needed to calculate KD values, even for complex interactions that generate multiple species, as shown above.
Refeyn’s TwoMP and TwoMP Auto mass photometers are both optimal for studying protein-protein interactions. In this experiment, data was generated using the TwoMP Auto, which offers automated pipetting for enhanced reproducibility.
Methodological details
Protein A, bovine & human IgG antibodies were purchased from Sigma-Aldrich
Mixtures of IgG and Protein A (1:1 molar ratio) were incubated for 1 hour at RT prior to measurement & transferred to a 96-well plate, which was loaded onto the TwoMP Auto mass photometer
Measurements were performed within 40 min using PBS as the buffer for droplet dilution find focus
The KD values were calculated as previously described3
References
1 Wu and Piszczek, Anal Biochem 2020
2 Wu and Piszczek, JoVE 2020
3 Soltermann et al., Angew Chem Int Ed 2020
4 Young, Biswas and Chen, Biophys J 2003
©2024 Refeyn Ltd
Unit 9, Trade City, Sandy Lane West, Oxford OX4 6FF, United Kingdom
For information on products, demos and ordering, write to info@refeyn.com
Refeyn is a registered trademark of Refeyn Ltd.
refeyn.com @refeynit Refeyn Refeyn
